This summer, I worked on incorporating a new source of DNA sequence annotations to Ginkgo’s sequence analysis software. During the design phase of the design-build-test cycle, biologists add annotations to their DNA constructs, or the engineered piece of DNA designed to enter the target organism. Even more specifically, during the design phase biologists annotate DNA design units, which are the building blocks comprising the DNA construct. Such annotations directly contribute to our goal at Ginkgo of making biology easier to engineer; by labeling subsequences of DNA with their functionality or any additional details through our DNA design tool, biologists can easily reuse DNA parts and leverage other biologists’ previously constructed DNA parts. When biologists think they have successfully inserted their DNA construct into the target organism, they input the DNA construct from the organism for sequencing, or reading the genetic code in their target organism. After sequencing, users visualize their sequencing data through the NGS sequence viewer. However, the NGS sequence viewer did not previously display the DNA design unit annotations users had created during the design stage.
For my summer project, I worked on integrating users’ design unit annotations from our DNA design tool into our sequence viewer for a more seamless user experience. By visualizing annotations with increased granularity in the sequence viewer, this project aims to increase the efficiency of biologists’ sequence validation, encourage high-quality annotations, and work to connect software tools across the design-build-test cycle of synthetic biology.
Interning at Ginkgo this summer has taught me a wealth of technical knowledge. I had the opportunity to work on a full-stack project, gaining exposure to Ginkgo’s tech stack as well as good software engineering practices. For the back-end portion of my project, I worked in a Python/Django framework to generate BED files for each annotation source. To communicate with the backend, I worked with GraphQL, learning the ins and outs of resolvers, queries and mutations. I learned how to use Docker networking to communicate between two different containers during local development. Finally, I worked in a Javascript/React frontend to visualize the two annotation types in our sequence viewer, learning about how to effectively organize React components into higher-level components and the intricacies of the React lifecycle.
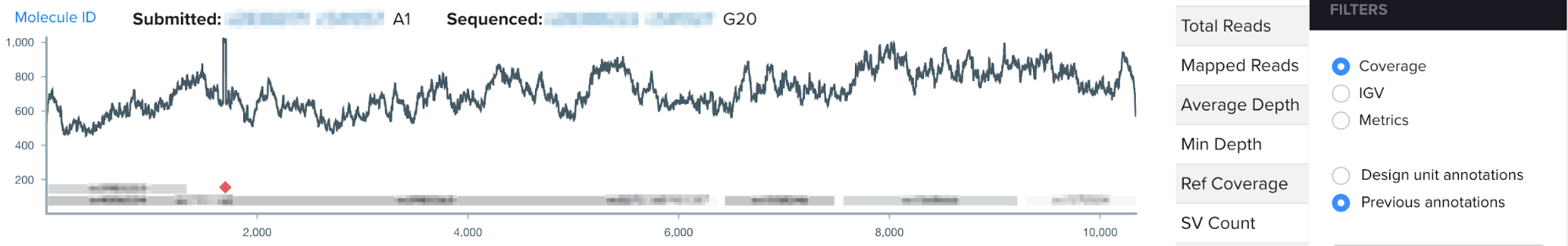
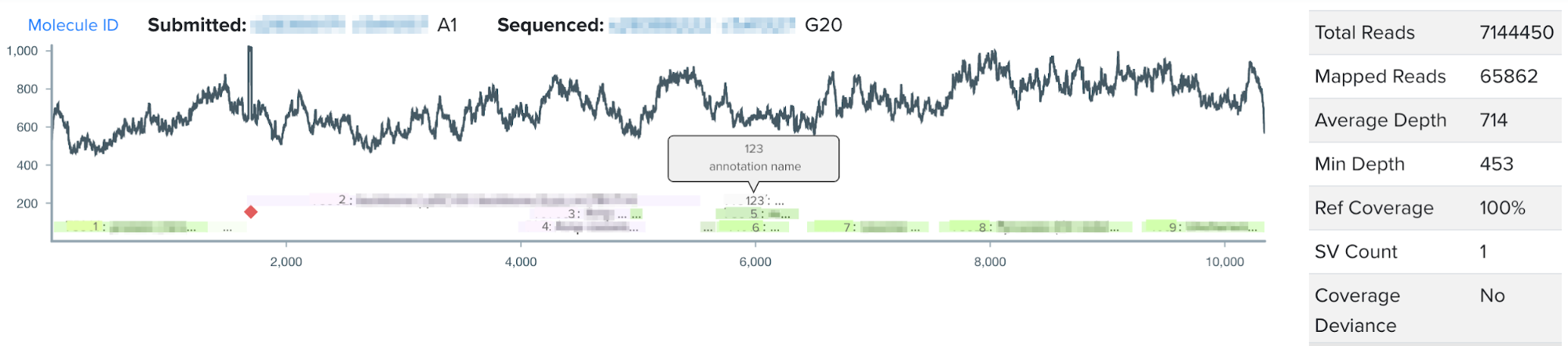
One of my favorite parts of my internship with Ginkgo was its structure, as I got to complete an end-to-end design process throughout the summer. I began the internship by interviewing biologists to identify the key pain points for users with the sequence viewer. I had the opportunity to collaborate and learn from UX designers, product managers, and other members of the software team to develop the vision for the feature, and achieve design consensus through a design review process. I also learned the value of using feature flags; by only deploying to specific users, we were able to gather valuable feedback on the feature and incorporate quick fixes before deploying to all users. To cap off the end-to-end design process, I led a training session for organism engineers to encourage use of the DNA design unit annotations in NGS viewer, and generate ideas for future versions of the feature.
Despite my internship being remote, Ginkgo made sure to enrich interns’ experience by exposing us to completely different areas of the company that we wouldn’t have otherwise interacted with. Within the Digital Tech team, we went to virtual lunches to meet with tech leads on teams different from our own, and participated in virtual hangouts with members of other digital tech teams. My mentor put on weekly leadership lunches for all interns across Ginkgo, where we got to hear from founders, from the Head of Codebase (the head of the program teams), and from Directors of Commercial Operations. Working at Ginkgo this summer has pushed me to grow both technically and professionally, and I could not be more grateful to my team for this opportunity!
(Feature photo by Ramakant Sharda on Unsplash)
